Experiments reported by Alex N. Nguyen Ba et al. in a paper titled “High-resolution lineage tracking reveals traveling waves of adaptation in laboratory yeast” in Nature.
The next group is doing evolution and ecology, and an undergraduate student came to do a project during the summer vacation. In such a short time, we can only use the existing data to write an article. This data comes from a paper titled "High-resolution lineage tracking reveals traveling wave of adaptation in laboratory yeast" by Alex N. Nguyen Ba et al. in Nature.
The experiment reported in the paper is briefly as follows:
- Insert a unique DNA sequence (the so-called barcode) into the genome of each yeast at the initial moment. After reproduction, yeast groups with the same barcode are considered to be the same "species";
- Allow this group of yeast to reproduce for 100 generations. During this period, monitor the proportion of yeast with different barcodes in the entire group and their changes, as well as their fitness to the environment;
- After 100 generations, insert new and different DNA sequences next to the original barcodes (because after evolution, many of the previous barcodes have been squeezed to a negligible proportion, and some of them occupy too large a proportion, and their offspring may have undergone new differentiation);
- Jump to step 2...
Barcode insertion
Barcode insertion uses enzyme-catalyzed gene recombination. The whole system is called Cre-Lox
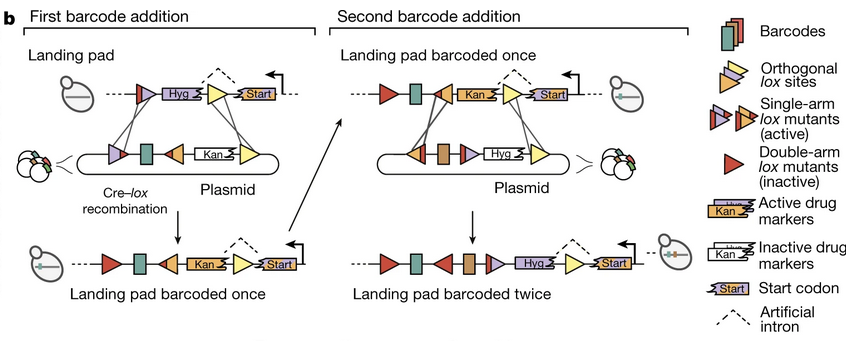
Lox is a gene, and there are many different types. Each type has a sequence called a spacer with uncertain content in the middle, and the sequences on both sides of the spacer with determined content are called arms; different specific sequences of the arms define different Lox types. The schematic diagram of the paper is represented by triangles.
Cre is a recombinase. In the presence of Cre, two DNAs with the same Lox gene that are close to each other have a certain probability of breaking at the Lox site, then exchanging halves and reconnecting. This exchange and reconnection is called gene recombination.
This experiment was performed on the diploid of a common budding yeast (Saccharomyces cerevisiae*), in which the position of the HO gene has been modified to become a promoter, a Cre gene, two different Lox fragments, and a selection marker sandwiched between the two Lox.
The barcode is located on an exogenous plasmid and is delivered into yeast cells through transformation. The barcode gene is flanked by two Lox fragments, with a selection marker on the right that is different from that in yeast, and a third Lox site on the right of the selection marker.
Genetic recombination in the Cre-Lox system is generally a reversible reaction, which is not conducive to the efficiency of barcode insertion. To solve this problem, the authors found that both arms of Lox can undergo genetic mutations. When only one arm mutates, the effect on recombination is small. When both arms of Lox are mutated, they will hardly recombine under Cre catalysis.
所以酵母基因组和质粒最左侧的 Lox 各有不同的一臂是突变型,条形码插入后两臂都成了突变型,就不会再通过重组离开酵母的基因组;
The lox next to the barcode is still a single-arm mutant, which is the starting position for the next barcode insertion. Using the same principle, different barcodes can be inserted multiple times.
After the insertion, we also want the plasmid to be excluded by the cell. This is achieved by placing the yeast cells in a 5-fluoroorotic acid (5-FOA) environment. Because the plasmid carries the Ura3 gene as a breeding marker, it can independently synthesize uracil (Uracil), and the two react to produce toxic 5-fluorouracil (Fluorouracil), so yeast that can survive in the selection environment no longer contain plasmids.
Cell culture, data reading and analysis
During the culture, 5 ml of yeast cell suspension was placed in a 15 ml glass test tube and kept shaking at 30 °C. According to previous papers, under this condition, the cells proliferated 10 generations per day. The authors diluted the cell suspension by times every day and took a certain volume of the suspension for gene sequencing; a new barcode was marked every 10 days, which was called 1 epoch; the experiment was carried out for a total of 10 epochs.
The purpose of gene sequencing is to count the number of yeast carrying each barcode at a certain moment. Because yeast is a single-cell organism, there is only one barcode in each cell, which means counting the number of molecules of various barcodes.
The method is to select primer sites upstream and downstream of the barcode, perform PCR amplification on the DNA molecules, make the number of DNA molecules significant enough, and reversely infer the original number of molecules based on the number of PCR cycles.
In order to prevent the PCR process from introducing additional errors into the experiment, a unique molecular marker (UMI) is added to each DNA. The method is to divide the PCR itself into two stages. The 5' end primer of the first stage PCR has a random sequence of 8-12 bases; the second stage PCR is carried out according to the normal configuration. The PCR products with the same molecular marker in the end can be regarded as originally coming from the same barcode molecule.
Relatively short gene sequences can be directly handed over to biotechnology companies (Illumina in this article) to read the sequence information, and the time and financial costs are affordable.
After reading the sequence information, the author wrote a regular expression-based algorithm using Python's regex library to perform analysis.
The core concept of evolution, fitness, is measured in this experiment by growing a group of yeast with a certain barcode together with an equal number of fluorescently labeled reference yeast. After the same period of time, the proportion of fluorescent cells in the total number of cells is observed to measure the ability of the yeast represented by the barcode to exclude the reference yeast. This method is called "competitive fitness assays".
Yeast sex
This section is not very useful for understanding the article, but I've already written anyway...
Yeast is a single-cell eukaryotic organism that can reproduce sexually and is generally diploid in the natural environment. During sexual reproduction, diploid cells first undergo meiosis to form haploid cells. Haploids have two possible mating types, MATα and MATa. The difference between the two is whether the content of the MAT gene is α or a. When the haploids of the two mating types fuse into a new diploid, sexual reproduction is completed.
Both mating types have a gene called HO, which encodes an endonuclease.
In the natural environment, HO can cause double-strand DNA breaks at the MAT site, triggering homologous recombination, thereby switching the mating type from MATa to MATα, or vice versa, thereby changing the sex of the haplotype. In this way, yeast can increase genetic diversity through sexual reproduction without mutation.
In the laboratory, researchers prefer that yeast do not change sex on their own, so they often knock out the HO gene or use the HO site as an insertion site for genetic engineering, such as the location where the barcode was inserted in this experiment. The gene shown in the upper left corner of Figure 1(b) was modified from the HO site.
See also: